|
By fbukolyi - Monday, October 2, 2006
|
|
See the screenshot, no tree icon in family C, H, P, V - though should be 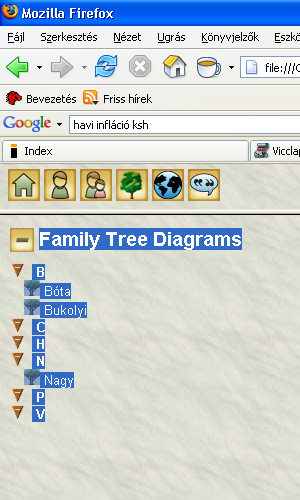
|
|
By GenoProSupport - Monday, October 2, 2006
|
|
Is this new to Beta18g, or it was there before in Beta 18f? Can you generate a sample file to http://familytrees.genopro.com, or send me your .gno file using the Send File command.
|
|
By fbukolyi - Monday, October 2, 2006
|
|
File sent to Genoprosupport. Some more info: when clicking on genomap Csirke and regenerate the report, this genomap also appears. When clicking on genomap Pomázi (regenerate), than this also will appear, however, H will remain still empty. Caching issue? 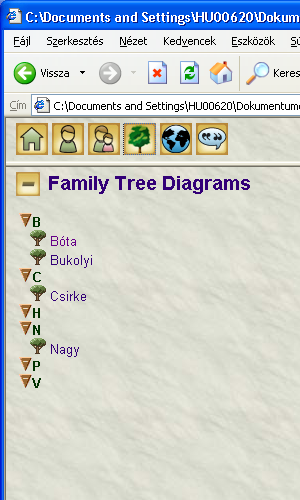 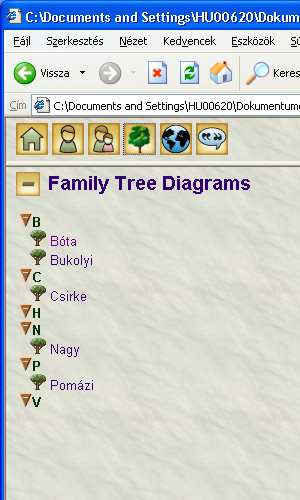
The generator settings are as follows: Options: nothing was set.Privacy: remove stray, no .gno, no .xml was set. Skin name English Narrative report.
|
|
By GenoProSupport - Tuesday, October 3, 2006
|
|
There is a problem indeed. I have contacted Ron about the issue.
|
|
By genome - Wednesday, October 4, 2006
|
|
It appears that the GenoMaps that do not appear in the list do not have the 'boundary_rect' attribute present. I guess these GenoMaps have not been updated for some time, as GenoPro would normally add the 'boundary_rect' attribute automatically. Applying a dummy update to one of the maps (e.g. add and then delete an individual) seems to cause GenoPro to add 'boundary_rect' to all the GenoMaps and fixes the problem.
|
|
By fbukolyi - Wednesday, October 4, 2006
|
You may right, Ron, as some maps were touched far ago. Maybe before that version, where
'boundary_rect' has been introduced  . Thanks . Thanks
|
|
By GenoProSupport - Wednesday, October 4, 2006
|
|
Ron (10/4/2006)
Applying a dummy update to one of the maps (e.g. add and then delete an individual) seems to cause GenoPro to add 'boundary_rect' to all the GenoMaps and fixes the problem.Just click on each GenoMap once and this is enough to generate a report with all the GenoMaps. Make sure you save to store the boundary_rect in the .gno file.
|