|
By Regislab - Sunday, March 24, 2019
|
Due to a lot of people appearing in my trees, I have created trees with external hyperlinks to avoid to have too big trees, I have just to click from link to link to follow the people in the trees. For example, one tree for my father with all their ancestors, one tree for my mother with all their ancestors, and so on. But when I wanted to run the Ancestors Report for myself from my own tree, the report doesn't consider the external hyperlinks, and just create the report from the tree where I started to run the report. How can I do this possible to get a report for all my direct ascendant, linked in all different trees ? Many thanks for any help. Gérard
|
|
By genome - Sunday, March 24, 2019
|
The GenoPro Report Generator does not follow external links and therefore a report skin cannot read details from another .gno.
I recommend holding all the data in one .gno, spread over different genomaps, or sets of genomaps. I have written a web-based app that allows your to create a .gno with a subset of the genomaps when you need to produce report on a particular group of genomaps. |
|
By Regislab - Sunday, March 24, 2019
|
Many thanks for this information about this, I will try it.  However I have just try to open the link but it seems not ok, I have attached the screen I have when I request the link 
|
|
By genome - Monday, March 25, 2019
|
I do not know what is wrong there. Could you please try again but after loading press F12 to display the devtools window and let me know the errors displayed in the console area.
|
|
By Regislab - Monday, March 25, 2019
|
|
Here is the printscreen you have requested, with doing F12, after using the link.
|
|
By genome - Monday, March 25, 2019
|
|
Should work now. It was only working in Chrome before. With a large .gno it can take some time to run. I think Chrome is quickest.
|
|
By Regislab - Monday, March 25, 2019
|
Yes and no  It works yes, and I can click on the "Continue" Button, but not on the next button concerning Drag and drop ...Here, attached the printscreen with F12 and Console again.
|
|
By genome - Monday, March 25, 2019
|
|
No errors shown. Try clicking on the actual text in the box not empty space. What browser are you using?
|
|
By Regislab - Monday, March 25, 2019
|
Ok, I have tried to click on the display box again, searching a position on the text where it should work and finally, I found one on the text, and it open it 
I use Mozilla Firefox.
I will try now to use correctly this tools. For the moment, I have not really understand how to get the report done with all my external hyperlinks, but I do not lose hope 
|
|
By genome - Tuesday, March 26, 2019
|
I have fixed the 'no click' issue now. 
|
|
By Regislab - Tuesday, March 26, 2019
|
I confirm, it is working 
|
|
By Regislab - Wednesday, March 27, 2019
|
I'm sorry to disturb you again about this, but I can't make this done. I have tried both possibilities, the first one, to drag and drop the .gno onto the box, which I need for the report, but it just save them or open them, any else. I have tried the other possibility, after clicking on the first box, and I have selected the .gno, than the GenoMaps I want to use. Same thing, it open them or save them. But I can't not select other .gno and needed GenoMap, at the same time. No Volume ist proposed to be selected, as you can see below :  In fact, I have not understand how to use correctly you tool. Is there somewhere an user guide for this ?
|
|
By genome - Wednesday, March 27, 2019
|
Perhaps you did not understand my original comment on this issue. I am suggesting the ALL your genomaps are held in one .gno, i.e. you do not use external hyperlinks but instead change them to internal hyperlinks.
But then when you wish to make a report on say just your father's ancestors then you can use this tool to produce a .gno containing the required subset of genomaps, not all of them. I hope this makes it clearer. You can preset the required genomap subsets, which I have called 'volumes' by setting values in the Report Title fields of the genomaps 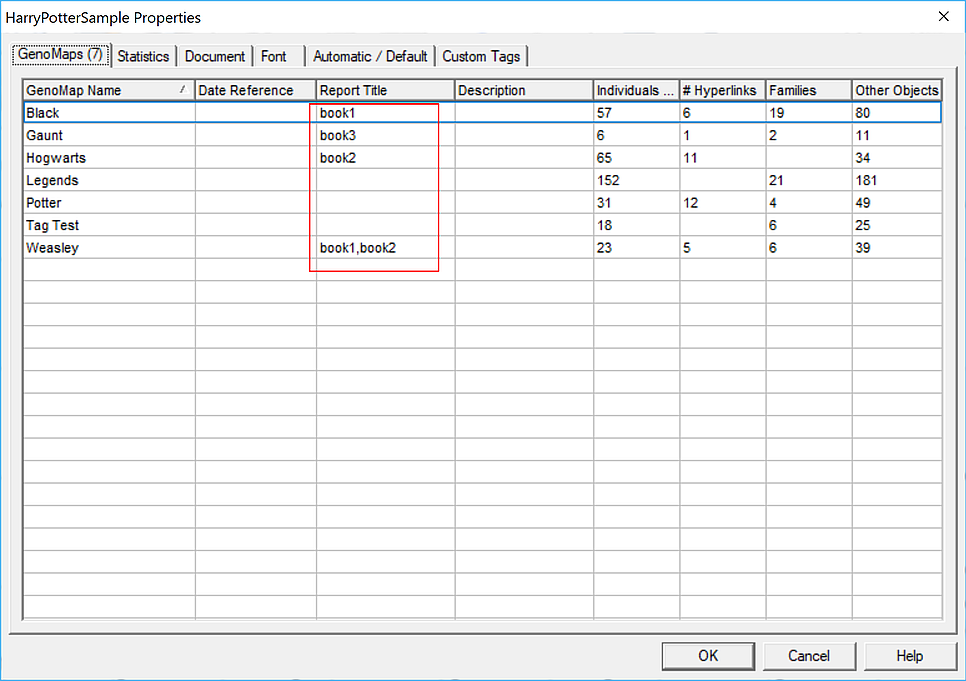 So here I have shown 'volumes' called book1, containing Black and Weasley genomaps, book2 with Hogwarts and Weasley and lastly book3 with just the Gaunt genomap. A list of any volumes set up in this way will appear in the second selection box of the subsetter tool.
|
|
By Regislab - Wednesday, March 27, 2019
|
I had understood the purpose of this tool, but I don't know how to use it correctly to create this .gno, which contains all datas, as you said, without the external hyperlinks, but with internal hyperlinks. It seems that your picture in the post is masking a part of your explanations, then down left the picture, we can just read "ook1" So I have tried and here what I got : what should I do now with this file called Data.xml ? |
|
By genome - Wednesday, March 27, 2019
|
The purpose of this tool is to create a smaller .gno from a larger one, this is what I mean by subset. It does not create a larger .gno from several smaller ones.
You will need to create the merged .gno yourself by say starting with the largest of your .gno files and then copying each genomap from each of the smaller .gno files and pasting into new genomaps in the largest one. It is also possible to merge .gno files (or actually their xml equivalents) providing they have different genomap names by using a text editor but this can get complicated and beyond the scope of this discussion. The advantage of one large .gno is that you do not have duplicate data (e.g. places, external linked individuals, sources)
|
|
By genome - Wednesday, March 27, 2019
|
rename Stammbaum-Balsiger.gno-2.zip to just Stammbaum-Balsiger.gno or save with that name to start with.
.gno files contain a compressed Data.xml file 
|
|
By Regislab - Wednesday, March 27, 2019
|
Okay all clear now 
I had effectively not understood this purpose but another one. If I will built a big tree, I will have to copy/paste more than 900 GenoMaps ! So I must decline this possibility. I can just hope that the reports will be able in the future to use also the external hyperlinks 
|
|
By genome - Wednesday, March 27, 2019
|
900 GenoMaps ! WOW!
Just out of interest how many .gno files and how many individuals do you have on those 900 genomaps?
|
|
By Regislab - Wednesday, March 27, 2019
|
|
16632 individuals :-) and 20 .gno
|
|
By Regislab - Wednesday, April 10, 2019
|
|
Can we hope to have the possibility that the report consider the external links too ?
|
|
By genome - Friday, April 12, 2019
|
I am afraid I cannot see that happening. Although GenoPro will open a .gno linked via an external hyperlink in a new window, from the Report Generator's viewpoint it remains a separate entity and this has to remain so as for example there is no way of telling what duplicates there are between two. Even the linked individuals can have different data so it is not possible to choose which one to use.
Also the GenoPro Report Generator is not likely to be enhanced as all effort is devoted to GenoProX. Therefore I stick to my original suggestion that .gno files are merged. This could possibly be automated to some extent. For example take the XML for each .gno and use search and replace in a text editor to ensure genomap names are unique by, say, adding a unique prefix for each .gno. This prefix could also be added as the Title tag of each GenoMap entry to serve as the 'volume' identifier, thus allowing the subsetter tool to reproduce the .gno if required. Then the XML files can be manually merged by copying all GenoMap objects to the GenoMaps section in target XML, Individual objects to Individuals section, Family to Families section, Place to Places section etc. For this to work then for technical reasons the first genomap in the source files must be empty, and so before editing the XML files you would need to, in each .gno, move everything from the first genomap to a new genomap, leaving the first empty. If this is not done then the objects in the first genomap of each .gno will all be placed in the first genomap of the target, as objects in the first genomap are not explicitly tagged with their owning genomap. Personally I would only use separate .gno for completely separate family trees, i.e. where there are no links.
|
|
By Regislab - Friday, April 12, 2019
|

|