|
|
|
|
Customers
FamilyTrees.GenoPro.com
GenoPro version: 3.1.0.1
Last Login: Wednesday, November 12, 2025
Posts: 264,
Visits: 1,760
|
I'm using some other tools/programs (eg Flourish Studio, FTDNA, DNA Painter, etc) that generate charts or graphics that I'd like to incorporate into my genomap.
While I can use png files for some of the sites, some of the charts are dynamic. Can I somehow block off an area on the genomap and plug in a webpage?
|
|
|
|
|
Administrators
Customers
Important Contributors
FamilyTrees.GenoPro.com
GenoPro version: 3.1.0.1
Last Login: Yesterday @ 8:18 PM
Posts: 3,466,
Visits: 26,882
|
You can use 'foreignobject' tag within a SVG tag to do this. e.g. Edit your genomaps.genomap?.htm file to add the tag after the highlight circle tag
<circle id="highlight" cx="0" cy="0" r="50" style="stroke-width:3px;fill:none;opacity:0.3;" visibility="hidden" pointer-events="none"> <animate attributeName="r" values="10;50" dur="5s" repeatCount="indefinite" /> <animate attributeName="stroke" values="fuchsia;fuchsia;white;fuchsia;fuchsia" dur="1s" repeatCount="indefinite" /> </circle><foreignobject width="400" height="300" x="100" y="100"><body><iframe src="https://www.wikipedia.org" width="400" height="300"/></body></foreignobject>
Choose width,, height, x, y, & src to suit. Some sites, e.g. www.google.com, are a bit picky about loading in an iframe but the is generally some way around it.
'lego audio video erro ergo disco' or "I read, I listen, I watch, I make mistakes, therefore I learn"
|
|
|
|
|
Customers
FamilyTrees.GenoPro.com
GenoPro version: 3.1.0.1
Last Login: Wednesday, November 12, 2025
Posts: 264,
Visits: 1,760
|
Forgive my ignorance, where do I find the genomaps.genomap?.htm file. I looked in the Common/Code folder of my GenoPro Skins folder but don't see it. I see genomap.htm and genomap.svg ... or am I looking in totally the wrong place? |
|
|
|
|
Administrators
Customers
Important Contributors
FamilyTrees.GenoPro.com
GenoPro version: 3.1.0.1
Last Login: Yesterday @ 8:18 PM
Posts: 3,466,
Visits: 26,882
|
Sorry should be genomaps/genomap?.htm (where ?=0,1,2,3 ...) and they are in your published report folder, not the skin templates. Choose the genomap in which you want to insert the web page.
If you edit the template file Common\Code\genomap.svg instead then the 'foreignobject' will appear in all genomaps.
'lego audio video erro ergo disco' or "I read, I listen, I watch, I make mistakes, therefore I learn"
|
|
|
|
|
Customers
FamilyTrees.GenoPro.com
GenoPro version: 3.1.0.1
Last Login: Wednesday, November 12, 2025
Posts: 264,
Visits: 1,760
|
I see my GenoPro Reports folder, but it only contains reports that I've run onto my computer.
The report on my computer is 5 years old, and I see all the genomaps are in the format I used then. Where would I find the genomaps/genomap?.htm files for reports that have only been uploaded to familytrees.genopro.com?
|
|
|
|
|
Administrators
Customers
Important Contributors
FamilyTrees.GenoPro.com
GenoPro version: 3.1.0.1
Last Login: Yesterday @ 8:18 PM
Posts: 3,466,
Visits: 26,882
|
They are in a similar place familytrees.genopro.com/userid/reportname/genomaps/genomap?.htm
Easiest way to see them is to open the report in Chrome, display the genomap required and then right click on it and choose 'view frame source' on the context menu. Then copy the text from the frame source window and paste into say Notepad on your PC and apply the edit to add the foreignobject markup. The amended file can be replaced on familytrees using https://genopro.com/help/ftp-client/I suggest you first create a label on the genomap with the size and position required and containing some unique text. You can then find this text in the frame source and it's rect tag and use its coordinates as a guide to create your foreignobject
'lego audio video erro ergo disco' or "I read, I listen, I watch, I make mistakes, therefore I learn"
|
|
|
|
|
Customers
FamilyTrees.GenoPro.com
GenoPro version: 3.1.0.1
Last Login: Wednesday, November 12, 2025
Posts: 264,
Visits: 1,760
|
Thank you! I will give this a try this weekend with some different web pages.
Not sure if this question should be covered in a separate thread: There is a javascript tool called Ideogram.js that allows you to display chromosomes and do a bunch of different things with them. Would there be a way to incorporate this tool into specific individual reports or onto a genomap? Here are some links to more information about Ideogram.js: https://eweitz.github.io/ideogram/https://github.com/eweitz/ideogramhttps://data.humancellatlas.org/analyze/visualization/ideogramAnother SiteJust some background FYI: - Currently, you send in a DNA sample to one of a handful of companies (eg, FTDNA, Ancestry, MyHeritage, etc)
- They test your sample, compare it to everyone else in their database, and give you an Ethnicity Estimate, and a list of DNA Matches (people you share a minimum amount of DNA with). Most companies give you a "Chromosome Browser" so you can see which specific segments are shared.
- You can download your Raw Data (a CSV file with ~700 K DNA locations) from any testing Company and upload it to almost every other Company or a Third Party Site to see other Ethnicity Estimates, other DNA Match Lists, or do different DNA visualizations. You can also just download just the Shared Segments to see what DNA you and your cousins inherited from your grandparents, great grandparents, etc.
- Currently, you either have to do a Screen Grab of the visualization, or the site generates a URL for you to link to. I sometimes use the Chrome Plug In GoFullPage to grab the very long visualizations.
- I'd like to be able to have that visualization capability right in GenoPro, possibly using Ideogram.js, instead of linking static pictures or external urls.
For example, if I've tested five 2nd Cousins, connected through 4 parents, 3 grandfathers, and 1 set of great grandparents, I'd like to have a diagram like the one below, except 4 layers deep, with 1, 3, 4, and 5 sets of chromosomes in each row, respectively. (Original here: Multiple Trio Ideogram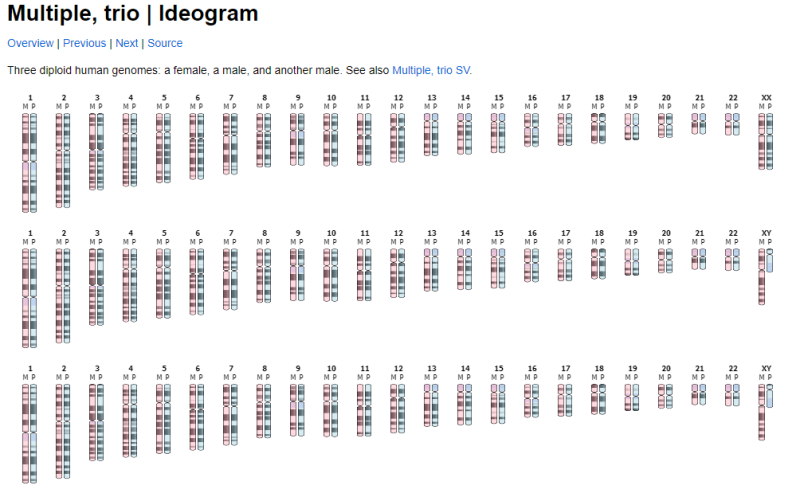 I'm fine if it's flipped on its side, or one chromosome at a time as well (Original link: Multiple Trio SV Ideogram ): 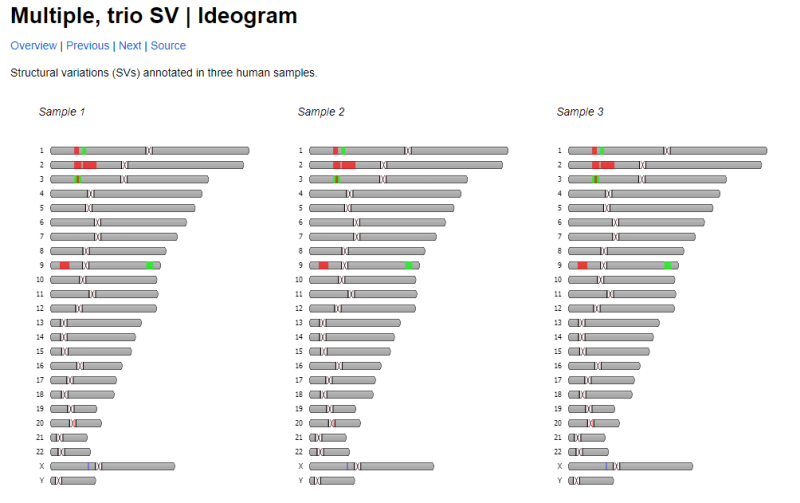 And then the multiple layers are linked to show which segments a pair of 2nd Cousins share, and how they passed up and down through the generations (If you click on the original web page, you'll see each bit of shared DNA gets highlighted when you hover over it: Advanced Homology Ideogram  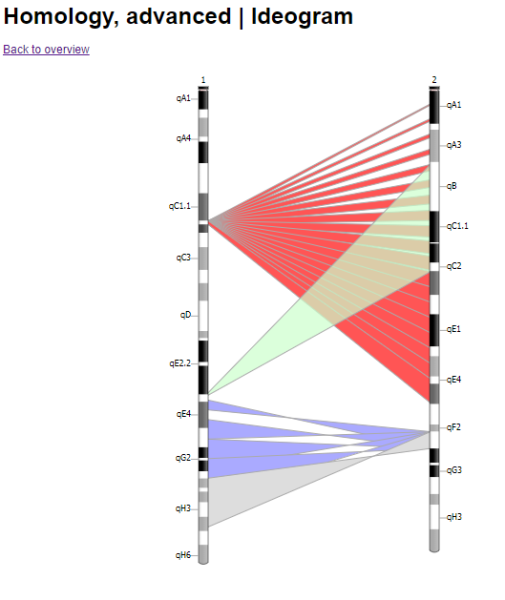 Here, I've tried doing that all by hand, but am not happy with the final results:  Edited: Friday, August 18, 2023 by
NiKo
Edited: Friday, August 18, 2023 by
NiKo
|